Abstract
The debate on issue of race in biology education requires a comprehensive reexamination. Traditionally, the idea that “we are all equal” has been emphasized, and that biology has rejected the concept of race. However, this perspective is problematic, as it contradicts current understandings about human genetic diversity, muddles debates about the legitimacy, utility, and possibility of categorizing humans, and perpetuates the invisibility of the inequalities faced by racialized individuals. This article advocates an alternative approach to addressing racial questions in biology classes by characterizing school science models that integrate concepts of genetics, ecology, and evolution. These models allow the discussion of certain common-sense ideas regarding racial questions while offering insights into exploring the nature of science in class. This approach encourages informed debates and a more profound understanding of the essence of scientific knowledge.
License
This is an open access article distributed under the Creative Commons Attribution License which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Article Type: Review Article
INTERDISCIP J ENV SCI ED, Volume 21, Issue 4, 2025, Article No: e2519
https://doi.org/10.29333/ijese/17441
Publication date: 19 Nov 2025
Article Views: 1301
Article Downloads: 478
Open Access HTML Content Download XML References How to cite this articleHTML Content
INTRODUCTION
Currently, addressing the issue of race in schools appears to be a pressing matter. Given the persistent inequalities faced by racialized people and the prevalence of hate crimes against them, it is worth asking what we can do in science education to rethink this issue in the classroom.
Traditionally, biology teaching has been one of the areas where this issue has been addressed. The traditional teaching approach focuses on highlighting the denunciation of scientific racism (Nascimento et al., 2019; Sánchez-Arteaga et al., 2013). However, students are rarely taught the underlying reasons behind biology’s rejection of the concept of race or the current understanding of human genetic diversity within biological populations (Donovan & Nehm, 2020; Duncan et al., 2024; Lee et al., 2021). This approach is also associated with the transmission of certain ideas by educators, such as the notion that all people are equal and that the concept of the human race does not exist, which in many cases leads to the avoidance of addressing this topic in the classroom (Aguilar, 2014; Pérez et al., 2025; Veeragoudar & Sullivan, 2022).
Presenting these ideas to students—or avoiding the topic altogether—can be problematic in several respects. From the perspective of biological models, claiming that “all people are equals” runs counter to the well-established fact of extensive genetic diversity among humans, even though this does not justify racial classification (Cela Conde & Ayala, 2014; Jobling et al., 2014; Rich Harris, 2010; Templeton, 2019). While biologists broadly agree that the traditional notion of race—which classifies individuals into groups such as Black, White, Asian, etc.—and its hierarchical structure lack scientific validity (Jobling et al., 2014; Lewontin, 1972), ongoing debates persist regarding the legitimacy, utility, and feasibility of categorizing humans into discrete groups based on genetic similarities (Barbujani & Colonna, 2010; Belbin et al., 2018; González Burchard et al., 2003; Novembre & Ramachandran, 2011; Rosenberg, 2011; Rosenberg et al., 2002; Serre & Pääbo, 2004). Ignoring this debate reflects a biased epistemological stance, presenting a rigid scientific perspective that fails to acknowledge the ongoing controversy within the field (Dunlop & Veneu, 2019; Fernández et al., 2002), and overlooking an important opportunity to engage with the contemporary role of biology in discussions about human races.
From the perspective of social science models, conveying these ideas—or avoiding the topic altogether—overlooks the complexities of ongoing scientific research. Social science approaches emphasize that people are not equal in social, political, or cultural terms. There are disparities in access to spaces and rights, even when such rights are proclaimed as universal. In many cases, race functions as a determining factor shaping this access (Fuentes, 2012; Identidad Marrón, 2021; Mullings, 2005). To assert that “we are all equal” or that “the human race does not exist” encourages students to view race as intangible, often leading to the conclusion that the solution to racism lies in being “colorblind” (Brookfield, 2019; Cooper Stoll, 2019; Graves Jr & Goodman, 2022; Veeragoudar & Sullivan, 2022). However, this perspective is problematic, particularly in light of disciplines such as anthropology, which argue that race—understood as a social rather than biological construct—plays a key role in shaping our lived experiences (Fuentes, 2020; Mullings, 2005). While thinking in terms of race can certainly fuel prejudice and discrimination, it also serves to illuminate the structural inequalities faced by individuals in areas such as healthcare, employment, and education. For example, Rutherford (2020) notes that during the COVID-19 pandemic, more Black people than white people died in the United States. These disparities cannot be fully understood without acknowledging race in its social dimension—and, by extension, racism. Ignoring this dimension would hinder genuine discussions about the inequalities faced by racialized groups and obstruct the development of targeted policies to redress the harm experienced by marginalized communities (Fuentes, 2012, 2020; Mullings, 2005). This stance reflects what anthropologist Peter Wade (2022) has called a “racism without race”.
Based on the aforementioned considerations, this paper proposes a novel approach to addressing the racial question in biology classrooms. In traditional approaches to biology education, models are often taught in ways that do not allow students to engage critically with the issue of race (Donovan & Nehm, 2020; Duncan et al., 2024). This work adopts a model-based perspective (Adúriz-Bravo, 2013; Giere, 1999, 2004). This perspective enables us to identify which biological models are useful for teaching this topic, delineating their boundaries and acknowledging their limitations in tackling a phenomenon as intricate as this one. The formulation of these models relies on the foundational frameworks of biology, the phenomena we aim to explore in the classroom, and the prevailing common-sense notions about human races that we want to discuss with the students.
The aim of this paper is to delineate a conceptual framework within biology that is both useful and meticulously updated, capable of addressing issues related to the genetic diversity of human populations, with a specific focus on the racial question.
To achieve this objective, the study follows what Grant and Booth (2009) refer to as a “critical review”. That is, a comprehensive examination of the literature that analyzes and synthesizes material, culminating in a synthesis of models. In this sense, we surveyed major academic publications in genetics, ecology, and evolutionary biology that present classical models; reviewed contemporary research literature on evolution and human diversity in the field of genomics; and engaged with key texts in the philosophy of biology that offer meta-analyses of these areas of biological research. It is important to note that we did not delve into texts from the social sciences, as the aim of this work is to elucidate which biological models contribute to thinking about the question of race. This task is particularly challenging because, unlike well-established classical models, definitions of concepts and models in these more recent theoretical developments remain relatively ambiguous and require further clarification.
MODELS FROM BIOLOGY TO ADDRESS THE RACE QUESTION
The proposal outlined below involves the integration of models from three distinct areas of biology:
-
Genetics,
-
Ecology, and
-
Evolution.
Acknowledging that the concept of “model” is polysemic—both in philosophy and in science education—this work adopts the model-based perspective developed by epistemologist Ronald Giere (1992, 1999, 2004). In this framework, a model is understood as an abstract, non-linguistic, socially constructed entity, recognized and accepted by a scientific community, which grants it a certain degree of “reality.” According to this perspective, models are not inherently true or false; rather, they are evaluated based on how well they correspond to specific aspects of the real world, in light of the purposes for which they were constructed. Consequently, models maintain a relationship of similarity (not isomorphism) with the phenomena they represent, signifying that any model will inevitably have limitations. These limitations stem from the fact that there will always be facets of the represented system that the model cannot fully encompass. This recognition also implies that there might not exist a singular model for the phenomenon of interest; rather, there could be a set of overlapping models that provide diverse perspectives, enabling a comprehensive understanding of the phenomenon from different angles (Giere, 1992, 1999, 2004; Passmore et al., 2014).
In this context, the models presented are designed to construct robust explanations grounded in the theoretical frameworks of biology regarding the genetic diversity of human populations. As mentioned in the preceding paragraph, these models will have specific limitations; they will allow us to elucidate certain aspects of the racial question, particularly those falling within the domain of biological understanding. In this regard, it is important to emphasize that biology can only shed light on certain dimensions of racial issues. As with any model, these insights are inherently limited, and a deeper understanding of the phenomenon will require the incorporation of additional models—such as those from the social sciences.
The models presented here can be classified as school science models, conceived through the didactic transposition of selected scientific models tailored for educational purposes (Adúriz-Bravo, 2013, 2019; Marzabal et al., 2024). From this perspective, it is expected that these school science models will be progressively reconstructed throughout students’ education, allowing them to increasingly approximate to the expert models (Carroll & Park, 2024; Marzabal et al., 2024).
In presenting these models, we describe the conceptual elements included in each one, which, during instruction, are interlinked, allowing students to collaboratively construct the intended model. Although some scholars argue that a model transcends its conceptual elements (Passmore et al., 2014), an emphasis on these elements is invaluable in the school context, particularly when designing teaching activities. This approach facilitates a more comprehensive understanding of the intricate biological concepts underpinning the racial question.
Figure 1 depicts a synthesis of pertinent school science models corresponding to each domain of biology, alongside specific concepts that emerge from the intersection of these three areas. An illustrative example of a didactic sequence grounded in these models can be found in Pérez and González Galli (2024, 2025).
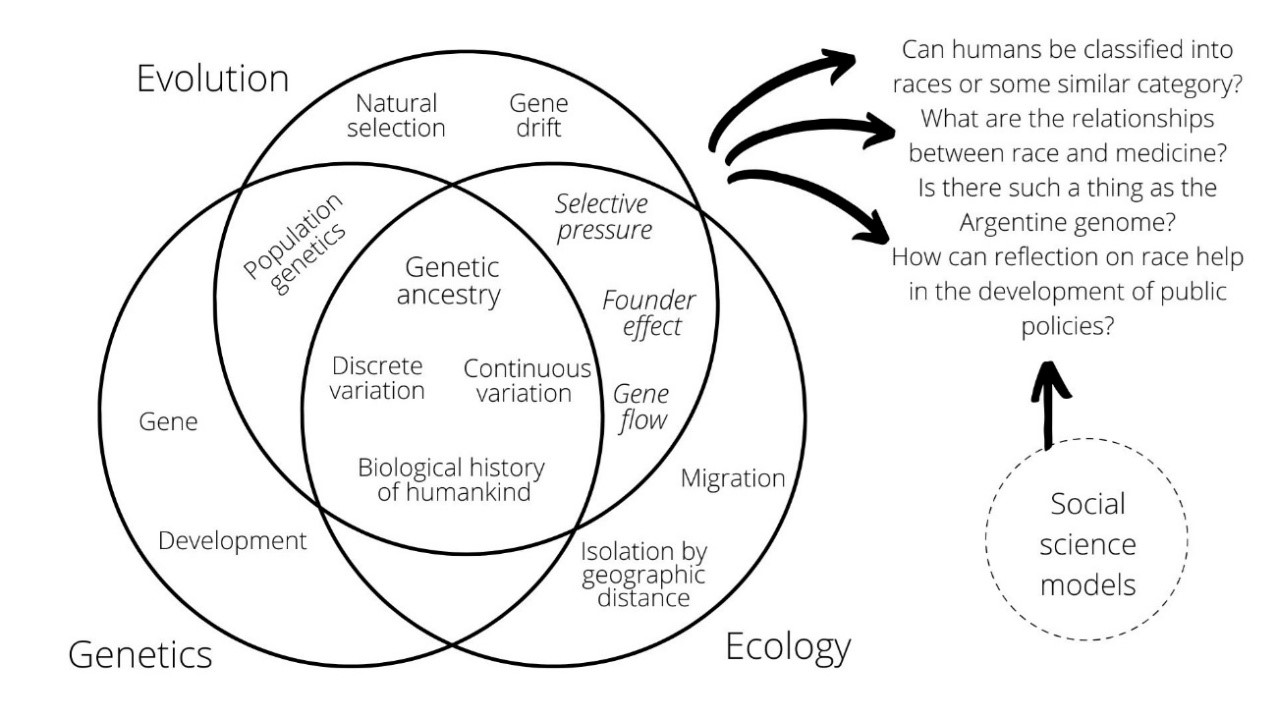
Introducing these models in mandatory education may enable students to develop a more complex perspective on the topic. This approach involves understanding current biological perspectives on the categorization of human populations, as well as recognizing the historical dimensions embedded in genetic ancestry. It should be noted, however, that a comprehensive understanding of this issue requires the inclusion of models from the social sciences—an aspect that will not be addressed in this work due to the specific disciplinary backgrounds of the authors and the aim of this article.
In the subsequent sections, each of these school science models will be delineated and their connections to the subject matter elucidated. Where applicable, their limitations will be emphasized, and the commonsense ideas about human races that enable their challenge will be outlined.
Gene Model
According to this model, genes are segments of DNA containing information necessary for constructing proteins that play a central role in forming an individual’s phenotypic traits. It is crucial to recognize that this genetic information contributes to the development of phenotypic traits but does not strictly determine them—this issue will be further explored in the following model. Variants of the same gene are referred to as alleles, and a gene with more than two variants is termed polymorphic. New alleles emerge through mutations of existing alleles (Jobling et al., 2014; Klug et al., 2013).
This basic gene model allows us to elucidate, among other aspects, that while all individuals possess genes related to skin pigmentation, there exist diverse variants of these genes among different people. For instance, concerning this trait, it is considered that approximately 15 genes contribute to the differences in individuals, encompassing those responsible for melanin quantity, melanosomes, or their arrangement in the epidermis (Barbujani & Colonna, 2010; Rutherford, 2020). Each of these genes harbors a multitude of alleles, leading to immense variability in skin tone, even within broad categories such as “white” or “black.”
Development Model
In this context, the term “development” does not refer to the gradual construction of body structures but rather to how various causal factors, encompassing both genetic and environmental elements, influence and interact during the formation of these structures. According to this model, an individual’s morphological, physiological, and behavioral traits (referred to as the phenotype) result from the interplay between their genes and the environment. This interaction includes stochastic processes occurring during development as well as environmental influences. Most human traits are polygenic, wherein each allele exerts a minor effect on the phenotype, and this effect is further modulated by environmental factors (Jobling et al., 2014; Klug et al., 2013; Rich Harris, 2010).
This model offers explanations for several phenomena in the world. For instance, regarding human height, it is believed that over 100 genes contribute to the variations in this trait. Even individuals with identical genetic patterns may not necessarily share the same height due to environmental factors such as nutrition, exposure to specific infections, or the level of oxygen available in their surroundings, all of which influence the development of this characteristic (Allen et al., 2010; Jobling et al., 2014). When an infant from a low-altitude region moves to a high-altitude area, various physiological mechanisms are triggered, impacting the individual’s height development (Relethford, 2012). Similar principles apply to traits such as skin pigmentation, predisposition to sports, or intelligence, which have historically been used to racialize people (Freeman & Herron, 2002; Rutherford, 2020).
Therefore, this model allows us to challenge the prevailing notion that human differences, including those historically used for racial categorization, are solely genetically determined. The developmental model presents an alternative explanation for this idea. Moreover, it prompts a reevaluation of the conventional belief in the direct correspondence between a single gene and a specific phenotype, a concept often reinforced by traditional approaches to Mendelian genetics in schools (Donovan et al., 2020, 2024).
Population Genetics Model
This model describes the genetic characteristics of a population, defined as a group of individuals belonging to the same species, residing in a specific area, and capable of reproducing with one another. Each individual within this population contributes a unique set of alleles, which are utilized to characterize the population based on their frequencies. Although all genes are present in all populations, what differs from one population to another is the number, type, and frequency of their alleles. Certain populations might possess alleles with higher frequencies than others, making them valuable as genetic markers. Furthermore, the concept of a population is perceived as an open genetic entity, signifying its ability to exchange alleles with other populations (Freeman & Herron, 2002; Rosenberg, 2011).
This model can be used to describe a wide range of phenomena related to the distribution of alleles around the globe. For instance, it elucidates the distribution of Alu variants, DNA segments lacking known function often utilized to establish parentage between individuals and populations (Bamshad & Olson, 2004). Similarly, it accounts for the distribution of gene variants facilitating the metabolism of alcohol or lactose (Evershed et al., 2022) and the distribution of blood groups (Fuentes, 2012). Furthermore, this model allows us to examine the fact that although human groups can be morphologically distinguished, research indicates substantial variation globally concerning different traits. In other words, racial distribution does not align with averages (Ehrlich, 2000).
However, while this model aptly describes the aforementioned phenomena, it possesses limitations when applied to human populations. The term “limitations” is used neutrally here and refers to aspects of the phenomenon not accounted for by the model (Giere, 1992, 1999, 2004). Specifically, the model does not consider additional factors influencing reproduction (and consequently allele distribution) such as social class, religion, among others (Relethford, 2012). These aspects are essential for understanding how societies are organized concerning race (Fuentes, 2020; Mullings, 2005).
Continuous Variation Model
According to this model, human genetic variation can be represented as a continuous gradient across geographical locations. This implies that there are no distinct boundaries between human populations, rendering the classical concept of human races biologically insignificant. This perspective draws heavily from a pivotal moment in the discourse on the racial question: Lewontin’s research in the 1970s (Lewontin, 1972). Lewontin argued that the diversity within populations far exceeds the diversity between populations or races, where the variation diminishes. Consequently, what have conventionally been termed human races exhibit much greater similarities among themselves, with the most significant variations occurring within each group among individuals. Genetic diversity within a population constitutes X% of the total variation, while the variation between populations accounts for (100-X) % of the total variation. The specific value of X varies across studies but generally falls within the range of 80 to 95% (Jobling et al., 2014; Rosenberg, 2011; Rosenberg et al., 2002).
Based on this understanding, the distribution of human genetic variation is best elucidated through a cline, defined as a gradient of allele frequencies or measures of genetic diversity across a geographic space along longitudinal or latitudinal ranges (Barbujani & Colonna, 2010; Cavalli-Sforza, 2000; Lewis et al., 2022; Pigliucci & Kaplan, 2003).
This model not only explains the original analyses conducted by Lewontin (1972) on blood groups based on traditional racial classifications but also subsequent analyses that arrived at similar conclusions (Jobling et al., 2014). Furthermore, it elucidates other patterns observed in genetic research, such as the finding that nearly 50% of alleles studied by biologists are present in all regions of the world (Rosenberg et al., 2002). Another phenomenon explicable through this model is the clinal nature of variation in skin pigmentation. The continuous variation in this trait precludes the construction of discrete categories with rigid boundaries, as stipulated by the classical concept of race (Relethford, 2012; Templeton, 2019).
Analyzing these phenomena provides an opportunity to engage students in critical discussions about the classical notion of race linked to visible phenotypes and the hierarchical structuring of these categories. Given that this model challenges the feasibility of racial grouping, it inherently questions the possibility of hierarchical classifications.
However, the continuous variation model has its limitations. Firstly, the specific clinal distribution observed depends on the polymorphisms used in its construction. For instance, in analyses involving mitochondrial DNA or the Y chromosome, values may differ due to less variation within populations and more variation between populations, possibly due to the greater influence of genetic drift (Jobling et al., 2014). This aspect will be further explored below. Secondly, methodological limitations exist in these studies, such as the univariate approach employed in these analyses. This approach considers only the allele averages for analysis and does not account for potential correlations between genes, which could contain pertinent information for defining groups (Rosenberg, 2011). This critique forms a central part of some challenges to Lewontin’s analysis, giving rise to discussions about the “Lewontin Fallacy” (Edwards, 2003; Sesardic, 2010).
Discrete Variation Model
According to this model, in contrast to the previous one, humans can be categorized into clusters based on genetics, often aligning with specific geographic regions. Notably, these genetic clusters do not align with traditional racial categories (Bamshad & Olson, 2004; Barbujani & Belle, 2006; Cavalli-Sforza, 2000; Hunley et al., 2009; Jobling et al., 2014; Rosenberg et al., 2002, 2005; Rosenberg, 2011; Rutherford, 2020; Templeton, 2013, 2019). The formation of these clusters depends on factors such as sample size, the geographical extent of the sample, the chosen number of clusters for definition, and the specific polymorphisms studied. Contrary to common intuition, each individual may belong to multiple clusters simultaneously (Barbujani & Belle, 2006; Barbujani & Colonna, 2010; Cavalli-Sforza, 2000; Rosenberg et al., 2005; Serre & Paabo, 2004; Templeton, 2019).
Biologists employ various genetic variations and conduct “clustering analyses” based on the genetic similarities among individuals to create these groupings. Within each group, individuals exhibit higher genetic similarity with one another than with individuals from other groups. Additionally, each group displays distinctive allele frequencies for the genes studied, differing from the frequencies of the same variants in other groups. Each individual is assigned a “membership value” for each group, representing the likelihood of that individual belonging to the group (this is another strongly counterintuitive aspect of this model, something we will have to keep in mind when teaching). This value can also be interpreted as the proportion of their genome originating from that particular group historically (Jobling et al., 2014; Serre & Paabo, 2004). This counterintuitive aspect is essential to consider when teaching this concept. Further elaboration on this point will be provided below.
This model allows to discuss that race is often defined by individual traits, such as skin color or hair shape, and that these traits are frequently used to distinguish people reliably. Simultaneously, it enables conversations with students about the argument that there are no discernible races, and we all belong to the same race: The human race. Working with this model in the classroom offers educators a platform to facilitate reflect on the utility, validity, and feasibility of categorizing humans into discrete groups. Engaging with this concept prompts further contemplation about the role of human classification in contemporary societies (Fuentes, 2020; Mullings, 2005).
In relation to the utility, the discrete variation model allows us to depict the varying prevalence of specific diseases in different populations. Certain diseases, including prostate cancer, hemochromatosis, blood coagulation disorders like factor V Leiden, familial hypercholesterolemia, among others, have been observed to exhibit different incidence rates across populations (Belbin et al., 2018; Foster & Sharp, 2002; González Burchard et al., 2003; Race, Ethnicity, and Genetics Working Group, 2005; Reich, 2018; Yashon & Cummings, 2011). Consequently, there exist specific genetic markers for these diseases with disparate allele frequencies among distinct population groups.
Knowledge about these genetic markers holds significant potential for “precision medicine,” a program rooted in understanding individual patient genomes and how genes interact with the environment, affecting susceptibility to specific diseases. The aim of precision medicine is to pinpoint specific genes or variants influencing disease risk or responses to particular drugs in individuals from particular populations. This approach tailors medical treatments to the unique genetic makeup of each person (All of Us Research Program Genomics Investigators, 2024; Belbin et al., 2018; Bustamante et al., 2011; Dopazo et al., 2019; Foster & Sharp, 2002; González Burchard et al., 2003; Klug et al., 2013; Risch et al., 2002; Rutherford, 2020; The International HapMap Consortium, 2003; Weigmann, 2006).
Discussing the concept of precision medicine in the classroom allows exploration of a common solution proposed to address racism, namely being “colorblind” (ignoring race). However, as demonstrated by the examples provided and the anthropological perspective mentioned earlier, disregarding racial categories would not be beneficial or desirable, particularly in medical contexts, as it could negatively impact people’s health. A pertinent illustration of this challenge arises from critiques of the overrepresentation of genetic markers from European or North American groups at the expense of markers from Latin American groups in precision medicine research (All of Us Research Program Genomics Investigators, 2024; Belbin et al., 2018; Bustamante et al., 2011; Dopazo et al., 2019; González Burchard et al., 2003; Krainc & Fuentes, 2022). This disparity affects research outcomes and exacerbates health-related inequalities between European/North American countries and Latin America. Countries possessing a better understanding of markers associated with diseases prevalent within their populations (typically European or North American countries) are better equipped to address the health needs of their people. Some authors (Brothers et al., 2021; Mullings et al., 2021) argue that this situation reflects institutional racism, emphasizing the importance of considering race in medical research to challenge traditional studies and institutional racism. Conducting “neutral” or “color-blind” medical research would neither be equitable nor advantageous, failing to reduce disparities in disease risk or treatment efficacy between different groups (Foster & Sharp, 2002; González Burchard et al., 2003; Risch et al., 2002).
Addressing this phenomenon in classrooms also allows for the discussion of certain ideas that people hold regarding the nature of science. For example, ideas about science being objective and culturally neutral (Brown, 2019). The discussion regarding the ways in which people are grouped based on their genetics, and the goals of these groupings, can be elements to question that image of traditional science.
The discrete variation model has some methodological limitations. For example, it is reported that some of these investigations use as a sample individuals belonging to very specific populations or cultures (Yoruba, Ashkenazi Jews, etc.), excluding those populations with mixed ancestry. This methodological decision generates that discrete clusters unfailingly appear in the groupings, which establishes an apparent substructuring of the population (Cela Conde & Ayala, 2014; Serre & Paabo, 2004; Templeton, 2019). Furthermore, no rigorous statistical criterion has been found to choose which number of clusters best represents human genetic variability (Templeton, 2019).
Some scientists are concerned that this type of research will help revive the traditional concept of human races (López Beltrán et al., 2017; Serre & Paabo, 2004; Templeton, 2019; Wade, 2014). Indeed, one of the criticisms of “precision medicine” is that race could be interpreted as the cause of the disease, with social variables that influence the development of such disease - such as socioeconomic status, to name a few - being unknown (Cooper et al., 1999; López Beltrán et al., 2017; Race, Ethnicity, and Genetics Working Group, 2005). It should be noted that what these investigations detect is a correlation and with this, there is always the risk of inferring - without grounds - a causal relationship, which would tend to reduce the explanation to genetic factors ignoring the other factors. Indeed, it is crucial for students to grasp the limitations of the discrete variation model. By understanding these limitations, students can develop a more nuanced and critical perspective on how genetic research is conducted and how race is interpreted within the realm of science. Acknowledging the complexity and constraints of these models fosters a more sophisticated understanding of the intricate relationship between genetics, race, and social factors, encouraging students to question assumptions and think critically about the information presented to them.
Migration Model
This model contributes to explaining the continuous genetic variation observed in the human population. It aids in understanding that individuals can belong to multiple clusters when genetic clustering occurs (Pigliucci & Kaplan, 2003; Rutherford, 2020; Templeton, 2019).
In genetic terms, migration refers to the movement of alleles from one population to another. Gene flow facilitates the transfer of alleles from one population to the allele pool of another. During this process, the divergence between populations decreases, leading to homogenization of allele frequencies. Consequently, the boundaries between populations blur, creating a continuum of variation (The 1000 Genomes Project Consortium, 2012; Freeman & Herron, 2002; Jobling et al., 2014; Pigliucci & Kaplan, 2003; Relethford, 2012; Rutherford, 2020; Templeton, 2019).
Throughout human evolution, numerous migration events occurred, including approximately 100 admixture processes within the last 4000 years (Hellenthal et al., 2014). There was even gene flow between different Homo species, including Denisovans and Neanderthals (Green et al., 2010; Novembre & Ramachandran, 2011; Reich, 2018; Sankararaman et al., 2014).
This model explains various phenomena, such as changes in AB0 allele frequencies over time within specific populations and historical admixtures among Europeans, Amerindians, and Africans (Avena et al., 2012; Belbin et al., 2018; Di Fabio Rocca et al., 2018; Luisi et al., 2020; Novembre & Ramachandran, 2011).
While the migration model challenges assumptions of racial purity, it has its limitations. For example, it does not account for several factors characterizing human migration, such as age, migrant distance, or language. Even migration in humans is sexually structured as well (Jobling et al., 2014).
Model of Isolation by Geographic Distance
This model helps to explain the pattern of discrete genetic variation, namely, why humans can cluster based on their genetics and why these clusters generally correspond to specific geographic regions.
According to this model, vast geographic distances and natural barriers such as oceans, mountains, or deserts hinder populations from intermixing, thus impeding gene flow. These conditions lead to inbreeding within populations on one side of the barrier, increasing similarities among individuals within the same side and accentuating differences with individuals on the other side of the barrier. This process results in a patchy distribution pattern of genetic variability (Barbujani & Colonna, 2010; Freeman & Herron, 2002; González Burchard et al., 2003; Rosenberg, 2011; Templeton, 2013). In the human species, such barriers include the Sahara Desert, large oceans, or the Himalayas, which were seldom crossed by humans during their migrations (Cela Conde & Ayala, 2014).
Several facts can be explained by this model, including the correlation between genetic clusters and specific geographic regions separated by natural barriers (Bamshad & Olson, 2004; Barbujani & Belle, 2006; Hunley et al., 2009; Jobling et al., 2014; Rosenberg et al., 2002). For instance, in the study by Rosenberg et al. (2002), these clusters corresponded to sub-Saharan Africa, Europe, parts of the West Asian Himalayas, East Asian regions of the Himalayas, Oceania, and the Americas.
Additionally, this model accounts for the existence of “private alleles,” which are genetic variants exclusive to specific populations and not found in others. Africa, in particular, boasts the highest number of private alleles (Rosenberg et al., 2005).
Similar to the discrete variation model, the isolation by geographic distance model challenges the common misconception that all humans are genetically identical.
Natural Selection Model
The natural selection model explains the distribution of certain traits (e.g., skin pigmentation, hair type, face configuration, height) as adaptations to specific environments (Bamshad & Olson, 2004; Cavalli-Sforza, 2000; Cela Conde & Ayala, 2014; Freeman & Herron, 2002; Nehm & Ridgway 2011; Relethford, 2012; Rutherford, 2020). According to this model, individuals in a population exhibit variability in almost all their traits. Heritable variants arise from random mutations that are not causally associated with the specific phenotypic effects they will produce. Some of these variants confer organisms a higher probability of reproducing and surviving in a given environment. Organisms possessing advantageous variants related to environmental factors will experience higher reproductive rates. Over generations, populations tend to increase the frequency of these advantageous variants, a process known as natural selection.
Genes subject to selection, such as those influencing skin pigmentation, height, or face shape, do not signify belonging to a particular race but provide insights into adaptations linked to the specific environments in which populations have lived (Cavalli-Sforza, 2000). For instance, concerning skin pigmentation, there exists a significant correlation between melanin levels and average UV radiation. This correlation likely results from a selection process, as melanin protects against UV rays but impedes vitamin D synthesis when UV intensity is low. Individuals living at the same latitude tend to share a similar type of pigmentation. Consequently, if this criterion were used, clusters formed based on such pigmentation would encompass people with diverse origins (Barbujani & Colonna, 2010; Cela Conde & Ayala, 2014). Research indicates that skin color evolved independently multiple times, even in populations not genetically isolated from others. Similar pigmentations reflect convergent evolution, where different populations adapted to similar selective pressures due to low sunlight levels and diets lacking in vitamin D (Pigliucci & Kaplan, 2003).
As explained above, the natural selection model allows to argue in biology classes that races can be characterized by skin color or other adaptive phenotypic traits.
Gene Drift Model
This model explains the random changes in allele frequencies within a population, leading to a decrease in variability due to stochastic sampling processes. Consequently, there is a tendency toward population homogenization, which stands in contrast to the diversifying effect caused by migration processes. Gene drift has a more pronounced impact when the population size is small (Barbujani & Colonna, 2010; Cavalli-Sforza, 2000; Freeman & Herron, 2002; Jobling et al., 2014).
In human evolutionary history, a specific manifestation of gene drift is the “founder effect.” This phenomenon occurs when a new population originates from a limited number of individuals from another population, resulting in a loss of genetic variability. During the founder effect, gene flow decreases as the distance between subpopulations increases, leading to divergence (Barbujani & Colonna, 2010; Freeman & Herron, 2002).
On a global scale, some studies argue that during human migration out of Africa, serial founder effects took place (Cela Conde & Ayala, 2014; Novembre & Ramachandran, 2011). This model explains the high frequency of rare variants in a geographically structured manner, such as specific founder mutations of breast cancer in Mexico, Chile, and Colombia (Zavala et al., 2019) or founder mutations of a gene variant influencing Laron syndrome in regions of Brazil, Chile, and Ecuador (Belbin et al., 2018).
Model of the Biological History of Humankind
This model explains various aspects of human biological history, including migrations, chronology, and geography. One crucial aspect of this model pertains to the emergence of anatomically modern humans, which occurred in Africa approximately 200,000 years ago. Several lines of evidence support this hypothesis:
-
Africa harbors more allelic variation than any other region in the world;
-
Alleles found outside Africa are a subset of the African allele pool;
-
The reconstruction of the mitochondrial DNA family tree traces its origins to Africa;
-
Nucleotide differences between two Namibian Khoisans are greater than those between a European and an Asian;
-
Region-specific alleles are rare but more common in Africa than in other continents; and
-
The fossil record and archaeological remains corroborate this spatial and temporal pattern (Barbujani & Colonna, 2010; Cavalli-Sforza, 2000; Cela Conde & Ayala, 2014; Freeman & Herron, 2002; Hardy, 2008; Henn et al., 2012; Jobling et al., 2014; Race, Ethnicity, and Genetics Working Group, 2005; Rosenberg et al., 2005, 2011; Templeton, 2013, 2019).
Other evidence could be presented to students, but the selected cases are considered necessary to maintain consistency with the other models presented.
As previously mentioned, human evolutionary history has been marked by gene flow between populations (Cavalli-Sforza, 2000; Novembre & Ramachandran, 2011; Pigliucci & Kaplan, 2003; Rutherford, 2020; Templeton, 2013, 2019). The oldest Homo sapiens specimens are from the southernmost tip of South Africa, near Cape Town. The migration of modern humans from Africa to the Near and Middle East occurred around 100,000 years ago, involving gene flow with Neanderthals around 80,000 years ago, possibly in the Near East (Green et al., 2010). Subsequent migration to Central and East Asia, Oceania, and Europe took place between 60,000 and 35,000 years ago, with the colonization of the Americas occurring approximately 15,000 years ago through a route now known as Canada (Cela Conde & Ayala, 2014).
Due to the extensive history of migrations and gene flow among diverse populations, some authors suggest that the traditional tree-like representation of human history does not fully capture the complexity of these interactions. Ancient DNA studies support network models, indicating significant mixing and gene flow between human populations (Rutherford, 2020; Templeton, 2019).
However, it’s important to note that this biological history model has limitations. It does not account for social, cultural, and political factors in human history, which have also influenced biological processes such as reproduction, isolation, and gene flow.
Genetic Ancestry Model
This model delves into the historical genetic relationships between populations. In the realm of genetics, ancestors are individuals who have biological descendants, and ancestry provides information about these individuals and their genetic connections to present-day people (Mathieson & Scally, 2020; Rutherford, 2020).
Consistent with other models, our evolutionary journey as modern humans has been marked by gene flow and admixture. Consequently, human DNA represents a mixture originating from various populations. This implies the absence of pure ancestries; in other words, there are no evolutionary lineages entirely independent of one another. Instead, individuals exhibit varying degrees of genetic components from different populations (Cela Conde & Ayala, 2014; Di Fabio Rocca et al., 2018; Freeland, 2005; Hardy, 2008; Jobling et al., 2014; Mathieson & Scally, 2020; Reich, 2018; Rutherford, 2020; Templeton, 2013, 2019).
Ancestry can be understood at both the individual and population levels. Concerning individual ancestry, it is possible to infer an individual’s genetic ancestry if specific alleles distinguishing populations are known. Despite the genetic similarities between populations, certain genetic variants may be more prevalent in some populations than in others. The collection of data on these variants enables the identification, with a certain probability, of an individual’s ancestral populations. Concerning population ancestry, the proportion of a population’s ancestry can be calculated based on its “parental” populations. This can be achieved by computing allele frequencies across all involved populations. Each parental population contributes a specific proportion of alleles to the hybrid population (Lewis et al., 2022; Jobling et al., 2014; Rosenberg, 2011).
Genetic ancestry estimation relies on two primary methodologies. One involves analyzing nuclear DNA, which allows the examination of genetic variants inherited from both parents. The other method involves analyzing mitochondrial DNA, which traces genetic variants exclusively from the maternal line, or Y chromosome DNA, inherited solely from fathers to XY offspring. Analyzing genetic variants on the Y chromosome, mitochondrial DNA, or nuclear DNA enables the assessment of the major “parental” populations of the current population. Nuclear DNA analysis facilitates the evaluation of interbreeding processes between populations over time. In contrast, analyzing Y chromosome or mitochondrial DNA sequences, due to their specific transmission characteristics, allows the distinct inference of an individual’s paternal or maternal lineage origin (Freeland, 2005; Freeman & Herron, 2002; Relethford, 2012).
As previously mentioned, populations have undergone various admixture processes throughout history. Migration episodes typically exhibit a general pattern involving a differential contribution from different populations, as observed in the analysis of mitochondrial DNA or the Y chromosome. In the case of Latin America, this pattern is a result of the historical impact of European colonization (primarily men) and the forced transfer of Africans during the slave trade. Numerous studies demonstrate genetic diversity patterns, particularly evident in the admixture found at the nuclear DNA level (Avena et al., 2012; Di Fabio Rocca et al., 2018; Luisi et al., 2020; Novembre & Ramachandran, 2011). Furthermore, diversity in mitochondrial DNA and Y-chromosomal DNA is also significant, introducing another layer to this model. Research generally suggests that there are more genetic variants from Native American populations in mitochondrial DNA, while European populations contribute more genetic variants in Y chromosome DNA. This phenomenon is explained by the concept of “sex bias,” indicating a differential pattern of genetic transmission, where each lineage (maternal and paternal) contributes disproportionate amounts of DNA to the mix (Jobling et al., 2014; Reich, 2018).
This intricate model of genetic ancestry enables us to explore various phenomena with students. For instance, it helps explain the diverse percentages of African, European, and Native American ancestry in Latin American populations (Belbin et al., 2018; Jobling et al., 2014; Novembre & Ramachandran, 2011: Templeton, 2019). It also sheds light on recent research findings indicating that between 1 and 4% of the genome comes from Neanderthals (Green et al., 2010; Novembre & Ramachandran, 2011; Sankararaman et al., 2014) or Denisovans (Reich, 2018).
Based on the presented information, this model facilitates discussions about common ideas such as the concept of the purity of human races and the notion that we all belong to the same race: Human race. It challenges assumptions related to “proud nationalism” and ideas concerning the preservation of national identity (Reich, 2018; Silva et al., 2020).
However, the genetic ancestry model does have limitations. Firstly, the idea of genetic ancestry sometimes simplifies identity, overlooking individuals’ self-perceptions of their ancestry associated with family histories, which significantly shape personal identity (Foster & Sharp, 2002; Mathieson & Scally, 2020). Hence, identity encompasses much more than genetics. Secondly, another limitation arises from the categorization of ancestry based on certain geographical (Asians, Northern Europeans, Africans, etc.), geopolitical (Norwegian, Zambian), or cultural (Ashkenazi Jews, Apaches, Yorubas) categories. These categories occasionally evoke racial classifications as they refer to “populations” analogous to them. If the scope and limitations of these categories are not well understood, they can mask substantial biological heterogeneity. Some categories even represent a much broader group. For instance, the term “African ancestry” often relies on genetic variants analyzed primarily from Yorubas in Nigeria, which is problematic as it generalizes African ancestry based on a very specific ethnicity, assuming homogeneity within the category that doesn’t reflect actual diversity. Some researchers (Kampourakis & Peterson, 2023; Lewis et al., 2022) consider that using these categories to describe individual ancestry is unnecessary, considering that the goal is to trace the paths through the human family tree by which an individual has inherited DNA from specific ancestors.
CONCLUSIONS AND IMPLICATIONS IN THE FIELD OF BIOLOGY EDUCATION
The aim of this paper was to establish a comprehensive and up-to-date conceptual framework in biology for addressing issues related to human genetic diversity, and more specifically the concept of human races. In total, eleven scientific models suitable for addressing this topic in compulsory education were proposed. In Figure 1, these models are visually represented, along with some of their interconnections. Throughout the study, we have emphasized addressing common misconceptions, many of which are implicit in racist discourses, using these scientific models. This effort aims to clarify the content that should be taught in the school, with a clear objective of promoting critical thinking regarding racist discourses. This approach aligns with the work of other researchers such as Donovan et al. (2020, 2024), Duncan et al. (2024) and Silva et al. (2020).
Teaching all these models simultaneously is not recommended; instead, they can be introduced gradually and explored throughout compulsory schooling. From our perspective, the teaching of these models should be aligned with specific learning objectives and the didactic transposition carried out by teachers, as each model allows for the exploration of certain aspects of the racial question. Previous works (Pérez & González Galli, 2024, 2025; Pérez et al., 2025) have shown how, within the framework of model-based teaching, some of these models can be addressed in compulsory education. This approach does not aim to replace students’ conceptions with scientific models, but rather to enrich and gradually complexify their ideas, bringing them closer to disciplinary models.
From this standpoint, the goal is not, for instance, to replace the idea that “all people are equal” with the idea that “all people are diverse,” but to complicate and deepen the former notion. In some respects, we are (or should be) equal, while in many others we are undeniably diverse. Nor is the goal to have students accept or reject the concept of race uncritically, but rather to recognize that the categorizations used in biology may resemble traditional racial categories to some extent yet also appear to be useful in medical contexts. In this sense, the process of complexification becomes a tension between models—a tension that is likely to remain limited if we focus solely on biological models. Therefore, we argue that, in order for students to develop more comprehensive and sophisticated understandings of the phenomenon in question, it is necessary to incorporate models from the social sciences—models that are beyond the scope of this article. The lack of effective integration between biological knowledge and social perspectives is a longstanding concern raised by anthropologist Agustín Fuentes (2012). As researchers in biology education, we believe that disregarding biological models for explaining human diversity deprives individuals of a fundamental tool for understanding our history. This gap in knowledge leaves people vulnerable to adopting and spreading misconceptions, such as those we have highlighted.
As illustrated in Figure 1, identifying specific models from the social sciences will be essential to enriching the conversation around the questions raised about human races. Looking ahead, collaborative efforts between biologists, social scientists, and educators are envisioned to design instructional activities that seamlessly integrate these diverse perspectives. In doing so, educational environments can be created in which students not only grasp the biological foundations of human diversity but also appreciate the complex interplay of social, cultural, and historical factors that shape human identities. This holistic approach will empower future generations to navigate the complexities of a diverse world with empathy, respect, and a deep understanding of human experience.
Author contributions: GP & LGG: conceptualization, project administration, supervision, validation, writing – original draft, writing – review & editing. Both authors have agreed with the results and conclusions.
Acknowledgement: Authors gratefully acknowledge the support of the Universidad de Buenos Aires and his fight for the right to a free, quality public education in the face of the affronts of the governments in power.
Funding: This work was supported by the Universidad de Buenos Aires through the project (20020220400219BA) “Estudio de los procesos de aprendizaje y de enseñanza de la biología evolutiva y la genética con énfasis en los obstáculos epistemológicos, metacognición, modelización, analogías y el pensamiento crítico”.
Ethical statement: The authors stated that ethical approval was not required for this study, as it is a theoretical and conceptual work that does not involve human participants, personal data, experiments, or the collection of sensitive information.
AI statement: The authors stated that no generative artificial intelligence or AI-based tools were used.
Declaration of interest: The authors report there are no competing interests to declare.
Data sharing statement: Data supporting the findings and conclusions are available upon request from the corresponding author.
References
- Adúriz-Bravo, A. (2013). A ‘semantic’ view of scientific models for science education. Science & Education, 22(7), 1593-1611. https://doi.org/10.1007/s11191-011-9431-7
- Adúriz-Bravo, A. (2019). Semantic views on models: An appraisal for science education. In A. Upmeier zu Belzen, D. Krüger, & J. van Driel. (Eds.), Towards a competence-based view on models and modeling in science education (pp. 21-38). Springer. https://doi.org/10.1007/978-3-030-30255-9_2
- Aguilar, J. (2014). Creencias sobre el concepto de raza en profesionales de la educación en Baja California [Beliefs about the concept of race among education professionals in Baja California]. Alteridades, 24(48), 123-136.
- All of Us Research Program Genomics Investigators (2024). Genomic data in the All of Us Research Program. Nature, 627(8003), 340-346. https://doi.org/10.1038/s41586-023-06957-x
- Allen, H., Estrada, K., Lettre, G., Berndt, S., Weedon, M., Rivadeneira, F., Willer, C., Jackson, A. U., Vedantam, S., Raychaudhuri, S., Ferreira, T., Wood, A., Weyant, R., Segrè, A., Speliotes, E., Wheeler, E., Soranzo, N., Park, J.-H., Yang, J., … Hirschhorn, J. N. (2010). Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature, 467, 832-838. https://doi.org/10.1038/nature09410
- Avena, S., Via, M., Ziv, E., Pérez-Stable, E., Gignoux, C., Dejean, C., Huntsman, S., Torres-Mejía, G., Dutil, J., Matta, J., Beckman, K., González Burchard, E., Parolin, M., Goicoechea, A., Acreche, N., Boquet, M., Ríos, M., Fernández, V., Rey, J., … Fejerman, L. (2012). Heterogeneity in genetic admixture across different regions of Argentina. PloS one, 7(4), Article e34695. https://doi.org/10.1371/journal.pone.0034695
- Bamshad, M., & Olson, S. (2004). ¿Existen las razas? [Do races exist?]. Investigación y Ciencia, 329, 6-12.
- Barbujani, G., & Belle, E. (2006). Genomic boundaries between human populations. Human Heredity, 61(1), 15-21. https://doi.org/10.1159/000091832
- Barbujani, G., & Colonna, V. (2010). Human genome diversity: Frequently asked questions. Trends in Genetics, 26, 285-295. https://doi.org/10.1016/j.tig.2010.04.002
- Belbin, G., Nieves-Colón, M., Kenny, E., Moreno-Estrada, A., & Gignoux, C. (2018). Genetic diversity in populations across Latin America: Implications for population and medical genetic studies. Current Opinion in Genetics & Development, 53, 98-104. https://doi.org/10.1016/j.gde.2018.07.006
- Brothers, K., Bennett, R., & Cho, M. (2021). Taking an antiracist posture in scientific publications in human genetics and genomics. Genetics in Medicine, 23(6), 1004-1007. https://doi.org/10.1038/s41436-021-01109-w
- Brown, M. (2019). Is science really value free and objective? In K. McCain, & K. y Kampourakis (Eds.), What is scientific knowledge? An introduction to contemporary epistemology of science (p. 226-241). Routledge. https://doi.org/10.4324/9780203703809-15
- Brookfield, S. (2019). Teaching race: How to help students unmask and challenge racism. Jossey-Bass.
- Bustamante, C., De La Vega, F., & Burchard, E. G. (2011). Genomics for the world. Nature, 475(7355), 163-165. https://doi.org/10.1038/475163a
- Carroll, G., & Park, S. (2024). Towards expansive model-based teaching: A systematic synthesis of modelling pedagogies in science education literature. Studies in Science Education, 1-39. https://doi.org/10.1080/03057267.2024.2417157
- Cavalli-Sforza, L. (2000). Genes, pueblos y lenguas. Crítica.
- Cela Conde, C., & Ayala, F. (2014). Evolución humana. El camino de nuestra especie [Human evolution. The path of our species]. Alianza Editorial, S.A.
- Cooper, R., Rotimi, C., & Ward, R. (1999). Hipertensión en los afroamericanos [Hypertension in African Americans]. Investigación y Ciencia, 271, 14-21.
- Cooper Stoll, L. (2019). Should schools be colorblind? John Wiley & Sons.
- Di Fabio Rocca, F., Spina, S., Coirini, E., Gago, J., Patiño Rico, J., Dejean, D., & Avena, S. (2018). Mestizaje e identidad en Buenos Aires, Argentina. Experiencias desde la búsqueda individual de datos genéticos [Mestizaje and identity in Buenos Aires, Argentina. Experiences from the individual search for genetic data]. Anales de Antropología, 52(1), 165-177. https://doi.org/10.22201/iia.24486221e.2018.1.62655
- Donovan, B., & Nehm, R. (2020). Genetics and identity. Science & Education, 29, 1451-1458. https://doi.org/10.1007/s11191-020-00180-0
- Donovan, B., Weindling, M., & Lee, D. (2020). From basic to humane genomics literacy. How different types of genetics curricula could influence anti-essentialist understandings of race. Science & Education, 29, 1479-1511. https://doi.org/10.1007/s11191-020-00171-1
- Donovan, B., Weindling, M., Amemiya, J., Salazar, B., Lee, D., Syed, A., Stuhlsatz, M., & Snowden, J. (2024). Humane genomics education can reduce racism. Science, 383(6685), 818-822. https://doi.org/10.1126/science.adi7895
- Dopazo, H., Llera, A., Berenstein, M. & González-José, R. (2019). Genomas, enfermedades y medicina de precisión [Genomes, diseases and precision medicine]. Ciencia, Tecnología y Política, 2(2). https://doi.org/10.24215/26183188e019
- Duncan, R. G., Krishnamoorthy, R., Harms, U., Haskel-Ittah, M., Kampourakis, K., Gericke, N., Hammann, M., Jimenez-Aleixandre, M., Nehm, R., Reiss, M. & Yarden, A. (2024). The sociopolitical in human genetics education. Science, 383(6685), 826-828. https://doi.org/10.1126/science.adi8227
- Dunlop, L., & Veneu, F. (2019). Controversies in science: To teach or not to teach? Science & Education, 28, 689-710. https://doi.org/10.1007/s11191-019-00048-y
- Edwards, A. (2003). Human genetic diversity: Lewontin’s fallacy. BioEssays, 25(8), 798-801. https://doi.org/10.1002/bies.10315
- Ehrlich, P. (2000). Human natures: Genes, cultures, and the human prospect. Washington: Island Press.
- Evershed, R., Smith, G., Roffet-Salque, M., Timpson, A., Diekmann, Y., Lyon, M., Cramp, L., Casanova, E., Smyth, J., Whelton, H., Dunne, J., Brychova, V., Šoberl, L., Gerbault, P., Gillis, R., Heyd, V., Johnson, E., Kendall, I., Manning, K., … Thomas, M. G. (2022). Dairying, diseases and the evolution of lactase persistence in Europe. Nature, 608, 336-345. https://doi.org/10.1038/s41586-022-05010-7
- Fernández, I., Gil, D., Carrascosa, J., Cachapuz, A., & Praia, J. (2002). Visiones deformadas de la ciencia transmitidas por la enseñanza [Distorted views of science transmitted through teaching]. Enseñanza de las Ciencias, 20(3), 477-488. https://doi.org/10.5565/rev/ensciencias.3962
- Foster, M., & Sharp, R. (2002). Race, ethnicity, and genomics: Social classifications as proxies of biological heterogeneity. Genome Research, 12, 844-850. https://doi.org/10.1101/gr.99202
- Freeland, J. (2005). Molecular ecology. John Wiley & Sons Ltd.
- Freeman, S., & Herron, J. (2002). Análisis evolutivo [Evolutionary analysis]. Prentice Hall.
- Fuentes, A. (2012). Race, monogamy, and other lies they told you: Busting myths about human nature. University of California Press. https://doi.org/10.1525/9780520951679
- Fuentes, A. (2020). Biological anthropology’s critical engagement with genomics, evolution, race/racism, and ourselves: Opportunities and challenges to making a difference in the academy and the world. American Journal of Physical Anthropology, 175(2), 326-338. https://doi.org/10.1002/ajpa.24162
- Giere, R. (1992). La explicación de la ciencia. Un acercamiento cognoscitivo [The explanation of science. A cognitive approach]. Consejo Nacional de Ciencia y Tecnología.
- Giere, R. (1999). Using models to represent reality. In L. Magnani, N. Nersessian, & P. Thagard (Eds.), Model-based reasoning in scientific discovery (pp. 41-57). Springer. https://doi.org/10.1007/978-1-4615-4813-3_3
- Giere, R. (2004). How models are used to represent reality. Philosophy of Science, 71(5), 742-752. https://doi.org/10.1086/425063
- González Burchard, E., Ziv, E., Coyle, N., Gomez, S., Tang, H., Karter, A., Mountain, J., Pérez-Stable, E., Sheppard, D., & Risch, N. (2003). The importance of race and ethnic background in biomedical research and clinical practice. New England Journal of Medicine, 348(12), 1170-1175. https://doi.org/10.1056/nejmsb025007
- Grant, M., & Booth, A. (2009). A typology of reviews: An analysis of 14 review types and associated methodologies. Health Information & Libraries, 26(2), 91-108. https://doi.org/10.1111/j.1471-1842.2009.00848.x
- Graves Jr, J. L., & Goodman, A. H. (2022). Racism not race: Answers to frequently asked questions. Columbia University Press.
- Green, R., Krause, J., Briggs, A., Maricic, T., Stenzel, U., Kircher, M., Patterson, N., Li, H., Zhai, W., Fritz, M., Hansen, N., Durand, E., Malaspinas, A., Jensen, J., Marques-Bonet, T., Alkan, C., Prüfer, K., Meyer, M., Burbano, H., ... Pääbo, S. (2010). A draft sequence of the Neandertal genome. Science, 328 (5979), 710-722. https://doi.org/10.1126%2Fscience.1188021
- Hardy, J. (2008). Race, genetics, and medicine at a crossroads. Lancet, 372, 85-89. https://doi.org/10.1016/S0140-6736(08)61886-3
- Hellenthal, G., Busby, G., Band, G., Wilson, J., Capelli, C., Falush, D., & Myers, S. (2014). A genetic atlas of human admixture history. Science, 343(6172), 747-751.
- Henn, B., Cavalli-Sforza, L., & Feldman, M. (2012). The great human expansion. Proceedings of the National Academy of Sciences, 109(44), 17758-17764. https://doi.org/10.1073/pnas.1212380109
- Hunley, K. L., Healy, M. E., & Long, J. C. (2009). The global pattern of gene identity variation reveals a history of long‐range migrations, bottlenecks, and local mate exchange: Implications for biological race. American Journal of Physical Anthropology, 139(1), 35-46. https://doi.org/10.1002/ajpa.20932
- Identidad Marrón. (2021). Marrones escriben. Perspectivas antirracistas desde el sur global [Browns write. Anti-racist perspectives from the Global South]. Universidad Nacional de San Martin.
- Jobling, M., Hollox, E., Hurles, M., Kivisild, T., & Tyler-Smith, C. (2014). Human evolutionary genetics (2nd edition). Garland Science. https://doi.org/10.1201/9781317952268
- Kampourakis, K., & Peterson, E. (2023). The racist origins, racialist connotations, and purity assumptions of the concept of “admixture” in human evolutionary genetics. Genetics, 223(3), Article iyad002. https://doi.org/10.1093/genetics/iyad002
- Klug, W., Cummings, M., Spencer, C., & Palladino, M. (2013). Conceptos de genética [Genetics concepts] (10 ed). Pearson education.
- Krainc, T., & Fuentes, A. (2022). Genetic ancestry in precision medicine is reshaping the race debate. Proceedings of the National Academy of Sciences, 119(12), Article e2203033119. https://doi.org/10.1073/pnas.2203033119
- Lee, J., Aini, R., Sya’Bandari, Y., Rusmana, A., Ha, M., & Shin, S. (2021). Biological conceptualization of race. Science & Education, 30(2), 293-316. https://doi.org/10.1007/s11191-020-00178-8
- Lewis, A. C. F., Molina, S., Appelbaum, P., Dauda, B., Di Rienzo, A., Fuentes, A., Fullerton, S., Garrison, N., Ghosh, N., Hammonds, E., Jones, D., Kenny, E., Kraft, P., Lee, S., Mauro, M., Novembre, J., Panofsky, A., Sohail, M., Neale, B., & Allen, D. (2022). Getting genetic ancestry right for science and society. Science, 376(6590), 250-252. https://doi.org/10.1126/science.abm7530
- Lewontin, R. (1972). The apportionment of human diversity. In T. Dobzhansky (Ed.), Evolutionary Biology (pp. 381-398). Springer. https://doi.org/10.1007/978-1-4684-9063-3_14
- López Beltrán, C., Wade, P., Restrepo, E., & Ventura Santos, R. (2017). Genómica mestiza. Raza, nación y ciencia en Latinoamérica [Mestizo genomics. Race, nation, and science in Latin America]. Fondo de Cultura Económica.
- Luisi, P., García, A., Berros, J. M., Motti, J., Demarchi, D., Alfaro, E., Aquilano, E., Arguelles, C., Avena, S., Bailliet, G., Beltramo, J., Bravi, C., Cuello, C., Dejean, C., Dipierri, J., Jurado Medina, L., Lanata, J., Muzzio, M., Parolin, M., Pauro, M., Sepulveda, P., Rodríguez Golpe, D., Santos, M., Schwab, M., Silvero, N., Zubrzycki, J., Ramallo, V., & Dopazo, H. (2020). Fine-scale genomic analyses of admixed individuals reveal unrecognized genetic ancestry components in Argentina. PLoS ONE, 15(7), Article e0233808. https://doi.org/10.1371/journal.pone.0233808
- Marzabal, A., Merino, C., Soto M., & Cortés, A. (2024). Modeling-based science education. In A. Marzabal, & C. Merino (Eds.), Rethinking science education in Latin-America (pp. 245-261). Springer. https://doi.org/10.1007/978-3-031-52830-9_13
- Mathieson, I., & Scally, A. (2020). What is ancestry? PLoS Genetics, 16(3), Article e1008624. https://doi.org/10.1371/journal.pgen.1008624
- Mullings, L. (2005). Interrogating racism: Toward an antiracist anthropology. The Annual Review of Anthropology, 34, 667-693. https://doi.org/10.1146/annurev.anthro.32.061002.093435
- Mullings, L., Torres, J., Fuentes, A., Gravlee, C., Roberts, D., & Thayer, Z. (2021). The biology of racism. American Anthropologist, 123(3), 671-680. https://doi.org/10.1111/aman.13630
- Nascimento, L., Sepúlveda, C., El-Hani, C., & Arteaga, J. (2019). Princípios de planejamento de uma sequência didática sobre a racialização da anemia falciforme [Principles of planning a didactic sequence on the racialization of sickle cell anemia]. XII ENPEC, Anais, Natal, 1-8.
- Nehm, R., & Ridgway, J. (2011). What do experts and novices “See” in evolutionary problems? Evolution: Education and Outreach, 4, 666-679. https://doi.org/10.1007/s12052-011-0369-7
- Novembre, J., & Ramachandran, S. (2011). Perspectives on human population structure at the cusp of the sequencing era. Annual Review of Genomics and Human Genetics, 12, 245-274. https://doi.org/10.1146/annurev-genom-090810-183123
- Passmore, C., Gouvea, J. S., & Giere, R. (2014). Models in science and in learning science: Focusing scientific practice on sense-making. In R. Matthews (Ed.), International handbook of research in history, philosophy and science teaching (pp. 1171-1202). Springer. https://doi.org/10.1007/978-94-007-7654-8_36
- Pérez, G., & González Galli, L. (2024). Metacognitive reflections on essentialism during the learning of the relationship between biology and the human race. Metacognition Learning, 19, 1035-1064. https://doi.org/10.1007/s11409-024-09394-x
- Pérez, G. & González Galli, L. (2025). ¿Quiénes somos y de dónde venimos? Propuesta para abordar el concepto de ancestría genética en la enseñanza de la evolución humana [Who are we and where do we come from? A proposal to address the concept of genetic ancestry in the teaching of human evolution]. In A. Gómez Galindo & G. Peñaloza (Eds), Enseñar y aprender evolución: Perspectivas latinoamericanas (pp. 141-171). Grañén Porrua Grupo Editorial.
- Pérez, G., González Galli, L., Corral, G., Corbelli, J., Salatino, L., Alegre, C. & Cupo, B. (2025). Ideas sobre la cuestión racial en estudiantes de profesorado y docentes de biología de Argentina [Ideas about race among teacher trainees and biology teachers in Argentina]. Revista de Educación en Biología, 28(2), 1-16. https://doi.org/10.59524/2344-9225.v28.n2.48251
- Pigliucci, M., & Kaplan, J. (2003). On the concept of biological race and its applicability to humans. Philosophy of Science, 70(5), 1161-1172. https://doi.org/10.1086/377397
- Race, Ethnicity, and Genetics Working Group. (2005). The use of racial, ethnic, and ancestral categories in human genetics research. The American Journal of Human Genetics, 77(4), 519-532. https://doi.org/10.1086/491747
- Reich, D. (2018) Who we are and how we got here: Ancient DNA and the new science of the human past. Oxford University Press.
- Relethford, J. (2012). Human population genetics. Wiley-Blackwell. https://doi.org/10.1002/9781118181652
- Rich Harris, J. (2010). No two alike: Human nature and human individuality. W.W. Norton & Company.
- Risch, N., Burchard, E., Ziv, E., & Tang, H. (2002). Categorization of humans in biomedical research: Genes, race and disease. Genome Biology, 3(7), Article comment2007.1. https://doi.org/10.1186/gb-2002-3-7-comment2007
- Rosenberg, N., Pritchard, J., Weber, J., Cann, H., Kidd, K., Zhivotovsky, L., & Feldman, M. (2002). Genetic structure of human populations. Science, 298, 2381-2385. https://doi.org/10.1126/science.1078311
- Rosenberg, N., Mahajan, S., Ramachandran, S., Zhao, C., Pritchard, J., & Feldman, W. (2005). Clines, clusters, and the effect of study design on the inference of human population structure. PLoS Genet, 1(6), Article e70.
- Rosenberg, N. (2011). A population-genetic perspective on the similarities and differences among worldwide human populations. Human Biology, 83(6), 659-684. http://doi.org/10.3378/027.083.0601
- Rutherford, A. (2020). How to argue with a racist: What our genes do (and don’t) say about human difference. The Experiment.
- Sánchez-Arteaga, J., Sepúlveda, C., & El-Hani, C. N. (2013). Racismo científico, procesos de alterización y enseñanza de ciencias [Scientific racism, processes of othering and science education]. Magis, Revista Internacional de Investigación en Educación, 6(12), 55-67.
- Sankararaman, S., Mallick, S., Dannemann, M., Prüfer, K., Kelso, J., Pääbo, S., Patterson, N., & Reich, D. (2014). The genomic landscape of Neanderthal ancestry in present-day humans. Nature, 507(7492), 354-357. https://doi.org/10.1038/nature12961
- Serre, D., & Pääbo, S. (2004). Evidence for gradients of human genetic diversity within and among continents. Genome Research, 14(9), 1679-1685. https://doi.org/10.1101/gr.2529604
- Sesardic, N. (2010). Race: A social destruction of a biological concept. Biology & Philosophy, 25(2), 143-162. https://doi.org/10.1007/s10539-009-9193-7
- Silva, A., Gouvêa Silva, A., & Franco, F. (2020). Utilização de conceitos evolutivos como contraponto a manifestações xenofóbicas [Use of evolutionary concepts as a counterpoint to xenophobic manifestations]. Investigações em Ensino de Ciências, 25(3), 70-85. https://doi.org/10.22600/1518-8795.ienci2020v25n3p70
- Templeton, A. (2013). Biological races in humans. Studies in History and Philosophy of Biological and Biomedical Sciences, 44(3), 262-271. https://doi.org/10.1016/j.shpsc.2013.04.010
- Templeton, A. (2019). Human population genetics/genomics and society. Human Population Genetics and Genomics, 437-473. https://doi.org/10.1016/B978-0-12-386025-5.00014-2
- The International HapMap Consortium. (2003). The International HapMap Project. Nature, 426, 789-796. https://doi.org/10.1038/nature02168
- The 1000 Genomes Project Consortium. (2012). An integrated map of genetic variation from 1,092 human genomes. Nature, 491(7422), 56-65. https://doi.org/10.1038/nature11632
- Veeragoudar, S., & Sullivan, F. (2022). Equity-based CS case study: An approach to exploring white teachers’ conceptions of race and racism in a professional development setting. ACM Transactions on Computing Education, 22(3), Article 28. https://doi.org/10.1145/3487332
- Wade, P. (2014). Raza, ciencia, sociedad [Race, science, society]. Interdisciplina, 2(4), 35-62. https://doi.org/10.22201/ceiich.24485705e.2014.4.47204
- Wade, P. (2022). El concepto raza y la lucha contra el racismo [The concept of race and the fight against racism]. Estudios Sociologicos, 40, 163-192. https://doi.org/10.24201/es.2022v40.2071
- Weigmann, K. (2006). Racial medicine: Here to stay? The success of the International HapMap Project and other initiatives may help to overcome racial profiling in medicine, but old habits die hard. EMBO reports, 7(3), 246-249. https://doi.org/10.1038/sj.embor.7400654
- Yashon, R., & Cummings, M. (2011). Human genetics and society. Cengage Learning.
- Zavala, V., Serrano-Gomez, S., Dutil, J., & Fejerman, L. (2019). Genetic epidemiology of breast cancer in Latin America. Genes, 10(2), Article 153. https://doi.org/10.3390/genes10020153
How to cite this article
APA
Perez, G., & González Galli, L. (2025). A synthesis on school scientific models of biology for addressing the question of human races. Interdisciplinary Journal of Environmental and Science Education, 21(4), e2519. https://doi.org/10.29333/ijese/17441
Vancouver
Perez G, González Galli L. A synthesis on school scientific models of biology for addressing the question of human races. INTERDISCIP J ENV SCI ED. 2025;21(4):e2519. https://doi.org/10.29333/ijese/17441
AMA
Perez G, González Galli L. A synthesis on school scientific models of biology for addressing the question of human races. INTERDISCIP J ENV SCI ED. 2025;21(4), e2519. https://doi.org/10.29333/ijese/17441
Chicago
Perez, Gaston, and Leonardo González Galli. "A synthesis on school scientific models of biology for addressing the question of human races". Interdisciplinary Journal of Environmental and Science Education 2025 21 no. 4 (2025): e2519. https://doi.org/10.29333/ijese/17441
Harvard
Perez, G., and González Galli, L. (2025). A synthesis on school scientific models of biology for addressing the question of human races. Interdisciplinary Journal of Environmental and Science Education, 21(4), e2519. https://doi.org/10.29333/ijese/17441
MLA
Perez, Gaston et al. "A synthesis on school scientific models of biology for addressing the question of human races". Interdisciplinary Journal of Environmental and Science Education, vol. 21, no. 4, 2025, e2519. https://doi.org/10.29333/ijese/17441

 Full Text (PDF)
Full Text (PDF)